This provides instructions of how to collect SEC-MALS-SAXS data at the BioCAT beamline. Before coming, please ensure that you’ve read up on how to plan your experiment and that you’ve properly prepared your samples.
Contents
Before data collection
- Every buffer you use for SEC-MALS-SAXS should be filtered and degassed.
- In SEC-MALS-SAXS experiments you should reserver ~50 mL of buffer in a falcon tube. This will be used for cleaning the sample loop, diluting samples, etc. It is convenient to not have to extract this from the larger buffer bottles.
- Every sample should be spun down for 10 minutes just before measurement.
Changing buffer
To change buffer you must do two things. First, you need to equilibrate the column in the new buffer. Second, you need to change the coflow buffer.
IMPORTANT: Never change directly from a buffer with salt to a storage solution of 20% ethanol, or vice versa. This could cause salt to crash out of solution and damage equipment. Always include a water equilibration step between those types of solutions.
Changing buffer on the Agilent
IMPORTANT: Talk to a beamline scientist before changing buffer on your own. In most SEC-MALS-SAXS experiments buffer changes are done by a beamline scientist.
SEC-MALS-SAXS setup needs long equilibration (~6 h to overnight), which precludes too many buffer exchanges during a single run. Please plan accordingly, and discuss with a beamline scientist if you have questions.
Be sure to never change the flow rate by more than 0.1 mL/min every minute.
Be sure you purge the RI flow cell.
You will also need to change the coflow sheath buffer.
Changing coflow sheath buffer
IMPORTANT: A beamline scientist should train you on how to change buffer before you do it on your own.
Whenever the buffer is changed for a SEC-SAXS or SEC-MALS-SAXS experiment the coflow sheath buffer also need to be changed. To do so:
- In the experiment control software, stop the coflow by clicking the “Stop Coflow” button.
- Uncap the new buffer. Remove the coflow buffer tubing from the current buffer.
- Remove the pierced cap from the current buffer and place it on the new buffer.
If there are any drops of the old buffer on it, gently with the cap with a kimwipe.
- IMPORTANT: You may be using the same buffer bottle for the HPLC as the coflow sheath. If so, make sure to stop the HPLC before transferring the HPLC leads with the coflow leads.
- Over a waste beaker, rinse the tubing with water from a DI Water squirt bottle.
- Place the coflow buffer line in the new buffer.
- In the experiment control software, start the coflow by clicking the “Start Coflow” button.
The coflow sheath flow should be given ~10 minutes to equilibrate. If you are low on buffer or doing a SEC-MALS-SAXS equilibration you can can then stop the sheath flow while the rest of the system equilibrates. If you are doing a SEC-SAXS equilibration and have plenty of buffer, BioCAT recommends leaving the sheath flow running so that you can’t forget to start it for your experiment.
Loading Sample
IMPORTANT: A beamline scientist should train you on how to load sample before you do it on your own.
Immediately before loading a sample you should spin down the sample for 10 minutes.
The Agilent systems use the vial autosampler to load sample, rather than an injection port. This means that multiple samples can be prepared at once, and run in short succession without opening the hutch, if so desired.
- Load sample volume + 50 µL into an autosampler vial.
- Put the vial in the autosampler tray. Take note of the position in the tray
and the tray number.
- Typically the positions used are those closest to the autosampler door and farthest from the needle home position in tray 1. For example vial position A1.
Set up data collection
Setting exposure parameters
IMPORTANT: A beamline scientist should train you on how to set exposure parameters before you do it on your own.
To set exposure parameters in the BioCAT control software:
Make a new folder for your sample by clicking on the folder button. It will be contained within your top level directory (should match all other top level directory names, such as 20190430Hopkins for a user group with PI Hopkins on 04/30/2019). Name the folder consistent with the sample identification in the FPLC/HPLC method.
- The BioCAT default for a sample name is PI initials plus sample number (starting at 1 and incrementing with each sample, for example JH1 for the first sample of a group with PI with initials JH).
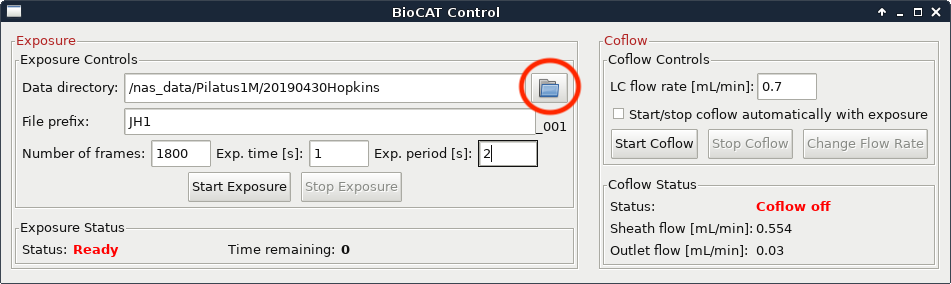
Change the filename to the new sample name. This should be consistent with the folder name and with the sample identification in the FPLC/HPLC methods.
- The BioCAT default for a sample name is PI initials plus sample number (starting at 1 and incrementing with each sample, for example JH1 for the first sample of a group with PI with initials JH).
Set the exposure time and exposure period appropriately. The defaults that most users will want to use are 1 s and 2 s for time and period respectively.
- Note: You will usually not need to change this. Check anyways just to to be sure.
Set the number of frames appropriately. The default most users will want to use is 1800. verify that frames*exposure period is equal to or greater than the run time of your FPLC/HPLC method.

Set the “LC Flow Rate” to the flow rate of your method. If coflow is on click the “Change Flow Rate” button.
- Note: You will usually not need to change this.

If coflow is off click the “Start Coflow” button.
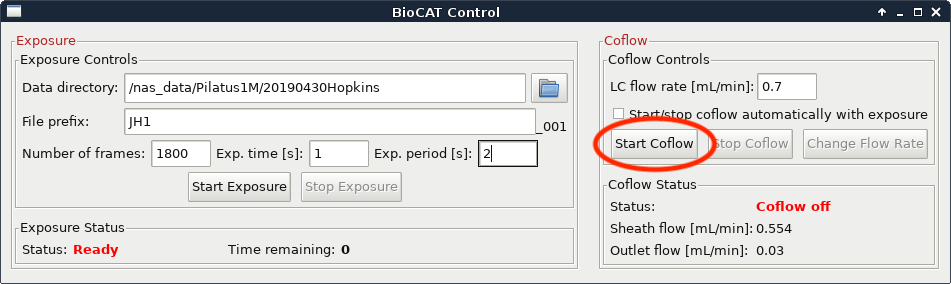
If you’re not sure what any of the above parameters should be, contact your beamline scientist.
Your exposure parameters are now set. You’re ready to start your data collection.
Start data collection
Starting data collection is now simple.
First
Then wait until a predetermined time and click the “Start Exposure” button. How long you wait depends on the column you are using, but generally speaking you should start the exposure just after the sample is injected. Talk to your beamline scientist for more guidance with your particular experiment.
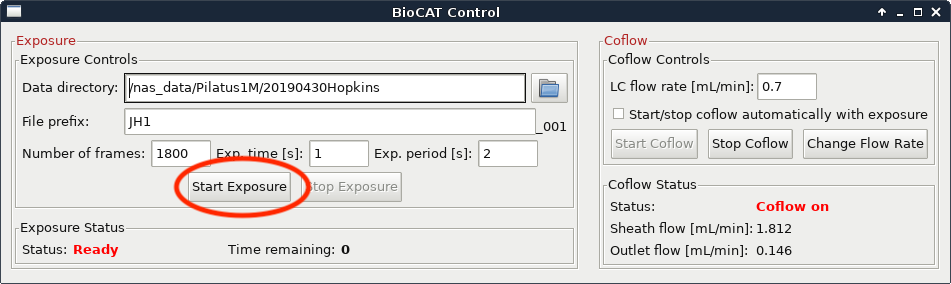
At this point you should also start on-line processing of the SAXS data.
Monitor the progress of the elution and the SAXS data to ensure nothing unexpected occurs during your run.
End data collection
The data collection will naturally end when your HPLC methods end and when your exposures end. If you are certain that you have collected all of the data (i.e. everything of interest has eluted and passed through the SAXS cell and the SAXS intensity has returned to baseline) you can end your data collection early. To do this, press the “Stop Exposure” button in the exposure control software.
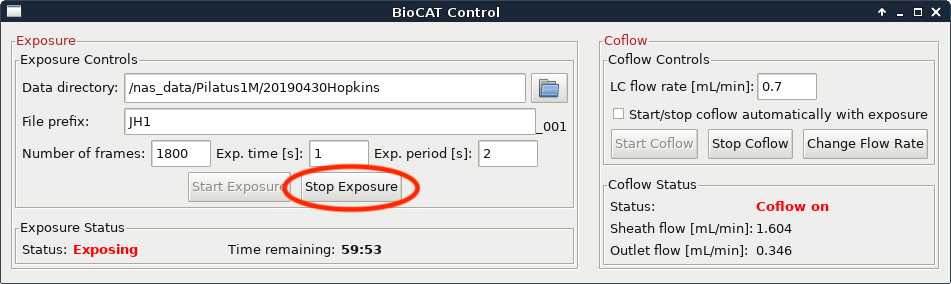
If everything has eluted from the injection (including any salt or other small molecules) you can also stop the HPLC method. Only do this if you are certain that everything has eluted, otherwise let it run the full 1-1.5 CV.
To do so
Optimizing your time
There are several things to keep in mind to help you optimize your time:
- Buffer changes on either instrument, but particularly the SEC-MALS-SAXS, take a lot of time. Optimize by combining samples into the same buffer as much as possible. Also make sure you know what experiments you’re doing in which buffer and do them all at once so you don’t have to re-equilibrate.
- If you are doing both SEC-SAXS and SEC-MALS-SAXS, you can do one or the other
while equilibrating the other system. A typical sequence might be:
- Equilibrate one or both of the SEC-MALS-SAXS systems overnight.
- In the morning at the start of your beamtime start to equilibrate the SEC-SAXS system.
- Collect data on one or both of the SEC-MALS-SAXS systems.
- Start those systems equilibrating.
- Switch to the SEC-SAXS system and run samples.
- Switch back to the SEC-MALS-SAXS systems.
- Groups with a lot of buffer changes can pre-equilibrate columns off-line on our preparative FPLC while running experiments on the AKTA.
- You should start spinning down your next sample with ~10-15 minutes left in your current run. This means starting to prepare any dilutions necessary as soon as you’ve started data collection on your current sample.
- If you’re sure all of the injection, including small molecules has eluted, you can stop your data collection early. Many users are able to stop data collection after 1 CV, and don’t need the entire 1.5 CV elution to clear the column.
- If you are using the SEC-MALS-SAXS instrument, once you have stopped the SAXS data collection you can load your next sample into the autosampler without waiting for the HPLC run to finish.
Older:
Collection Workflow
- Switching buffer/equilibration
- SEC-MALS-SAXS setup needs long equilibration (~6 h to overnight), which precludes too many buffer exchanges during a single run.
- Split buffer in half so both inlet A and B can pump buffer. Be prepared for enough buffer and bring them in two bottles in advance. 2L is usually sufficient.
- Change flow rate by 0.1 ml/min about every minute (after pressure levels out) as you stop the flow of one buffer and begin the flow of a new buffer.
- Once back up to 0.8-1 ml/min, equilibrate for at least 6 more hours.
- If equilibrating overnight, we can equilibrate at a lower speed overnight (0.2 ml/min) and ramp up to operational flow-speed 2 hours before the experiment). For 6 hr equilibration, ramp up to operational flow speed (0.8-1 ml/min) right at the beginning.
- Make sure you clean the flow cell for the Optilab - T-rEX in between buffers, especially if there’s more glycerol etc (anything that could change refractive index) - this requires the “purge” button on the LED panel to be switched on.
- ALSO: keep glycerol concentrations as low as possible (preferably <5%).
- Injecting sample and starting HPLC run
- Have samples ready (concentrate or dilute to appropriate concentration
and spin down 10-15 minutes) so that near the end of one run you can
- Have the auto-sampler inject your sample just as the next run is about to start
- Each run should be about done after ~30 min (may vary based on sample elution
profile and flow rate and usually signified by the integrated scattering
intensity coming back to baseline levels) - stop collecting SAXS data
then and you will have 5 min to put the new sample
in the sample tray
- If you think you will be running late, time can be extended
- To change time, right click in Quat. pump, select Method, change time
- Preparing your sample
- Samples are injected from vials. 250 - 350 µl will be injected, but fill vials to ~50 µl more than the injection volume (900 µl is the upper limit).
- Loading your sample
- At the beginning of each new sample set, set a program for that sample set
- In ASTRA (Wyatt’s MALS software):
- Create new sequence file, type number of samples and name them
- Set the sample property 1) name, 2) choose method (MALS-dRI-SAXS), 3) set time (18-23 mins depending on flow rate) for each sample
- In Chemstation (Agilent’s HPLC software):
- Use the sample entry window to select positions in sample tray and name them
- Set the method SEC_constflow for each sample
- Make sure the sample volume is appropriate and make necessary changes before starting the program.
- In ASTRA (Wyatt’s MALS software):
- For each sample, put the vial in the proper position in the tray for that sample.
- Watch the autosampler pick up the vial, aspirate the sample, and replace the vial
- Once the vial has been replaced and the robot moves out of the way, remove the vial and check that a reasonable amount of sample has been aspirated to ensure proper functioning of the autosampler. Begin station search (make sure capillary has been cleaned and is hooked back up to HPLC)
- At the beginning of each new sample set, set a program for that sample set
- Data from HPLC runs are saved in pre-determined folder (usually date followed by PI last name).
- Have samples ready (concentrate or dilute to appropriate concentration
and spin down 10-15 minutes) so that near the end of one run you can