The nucleic acid RNA plays an important role in protein synthesis in cells. However, noncoding RNAs also exist that are not converted into proteins, but still play important roles in many biological processes. RNA molecules aggregate into complex tertiary structures, producing globular forms stabilized by various interactions. Proteins, ligands, and other RNA molecules recognize tertiary folded RNAs and result in the biochemical pathways that affect all aspects of cellular metabolism. Using small-angle X-ray scattering (SAXS) at the APS, and other techniques, researchers investigated the unique folding behavior of RNA. They described how this occurs with the cooperation of folding intermediates of an RNA enzyme, ribozyme. The native structure of ribozyme was important for this coupling reaction, with small alterations in its architecture determining the entire folding pattern. This interaction took place early in the folding process, with the formation of structural themes, or motifs, being linked in near-native folding intermediates. Cooperativity was also found to require the orientation of the native helix. Tertiary interactions had little effect on the stability of the native state of the ribozyme. Understanding the results of this research will be important in guiding future studies to further evaluate the importance of RNA tertiary structure in biological systems.
RNA is one of the 2 types of nucleic acids found in all cells. Its main role is to carry out instructions for protein synthesis from DNA, the 2nd type of nucleic acid which stores the genetic information in cells. While messenger RNA represents the RNA that codes for protein synthesis, noncoding RNAs also exist that are not translated into protein. Noncoding RNAs are found widely in biology, having roles in the process of protein translation or gene regulation, for example. Some can also catalyze chemical reactions such as cutting and ligating RNA molecules, and these are known as ribozymes. It is now thought that noncoding RNAs code for even more biochemical functions than originally suspected. This range of involvement in cellular processes relates to the ability of RNA to serve as a carrier of genetic information and to adopt unique, complex three-dimensional structural folds that are critical to their function - creating sites that allow for chemical reactions or control gene expression.
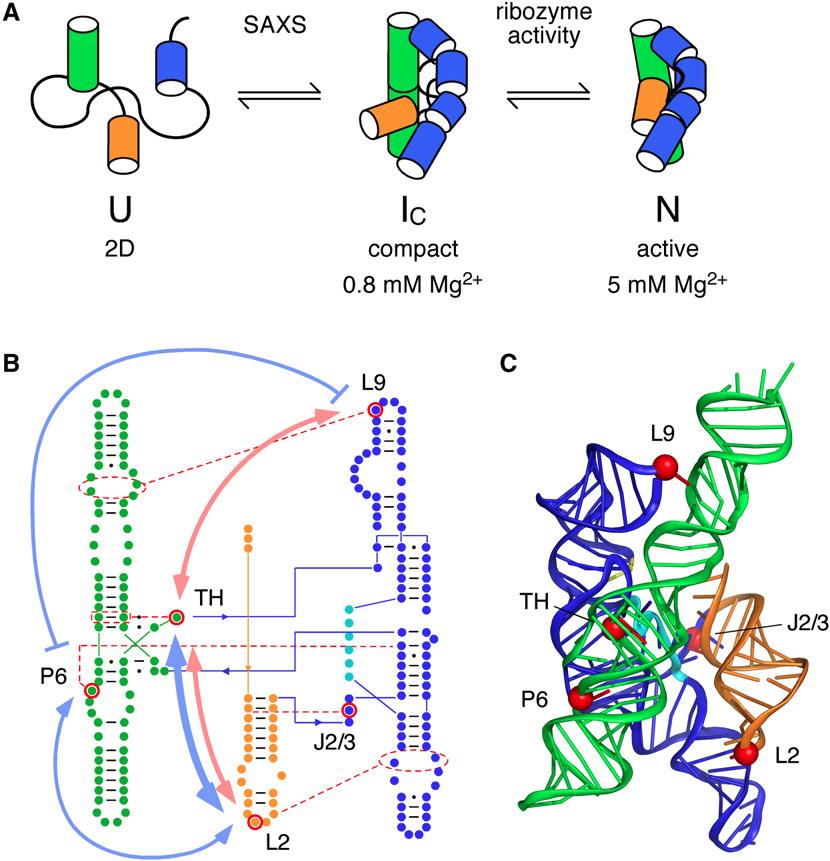
The tertiary structure of RNA refers to the three-dimensional arrangement of RNA building blocks that are held together via connections known as tertiary interactions. Although studies of noncoding RNAs have revealed the existence of structural themes known as tertiary motifs, as well as organizational principles, the exact mechanism by which these encode the self-assembly of unique three-dimensional RNA structures remains poorly understood. This study aimed to examine why RNAs fold so specifically in spite of the relatively small number of tertiary motifs.
Using SAXS at BioCAT ID18 at the APS, as well as other techniques, to measure changes in the folding energy landscape, the researchers showed that these tertiary interactions are highly related to the folding intermediates of ribozyme, an RNA enzyme ribozyme. They also showed that most tertiary interactions provide only small contributions to overall stability of the native state of the ribozyme. However, the formation of structural motifs is cooperatively linked in near-native folding intermediates, and this interaction depends on the native helix orientation. They demonstrated how this cooperativity occurs early in the RNA folding process. Coupling between tertiary structures in different areas of the RNA inhibits nonnative structures, while favoring the active RNA structure by increasing the free energy gap between the native state and the next most stable structure, thus simplifying the search for the native fold. The native structure of ribozyme is important for this coupling reaction, with small alterations in its architecture determining the entire folding pattern.
This study has provided important insights into the importance of these early interactions in the RNA folding process, and indicates that cooperativity in noncoding RNAs may have arisen as an evolutionary process due to natural selection of structures that favor formation of unique folds. The results of this study increase our knowledge of how tertiary interactions in RNA are related through its helix, and how they promote cooperative self-assembly. This work will help to guide further research into the components of tertiary RNA structure, and will ultimately further enhance understanding of important information about its biological functions.
Adapted from a Advanced Photon Source press release by Nicola Parry
See Reza Behrouzi, Joon Ho Roh, Duncan Kilburn, R.M. Briber, Sarah A. Woodson, “Cooperative Tertiary Interaction Network Guides RNA Folding,” Cell 149 (2), 348-357 (2012). DOI: 10.1016/j.cell.2012.01.057