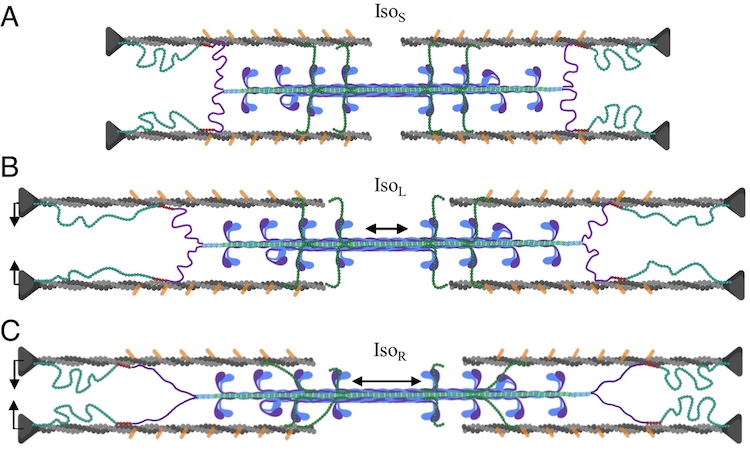
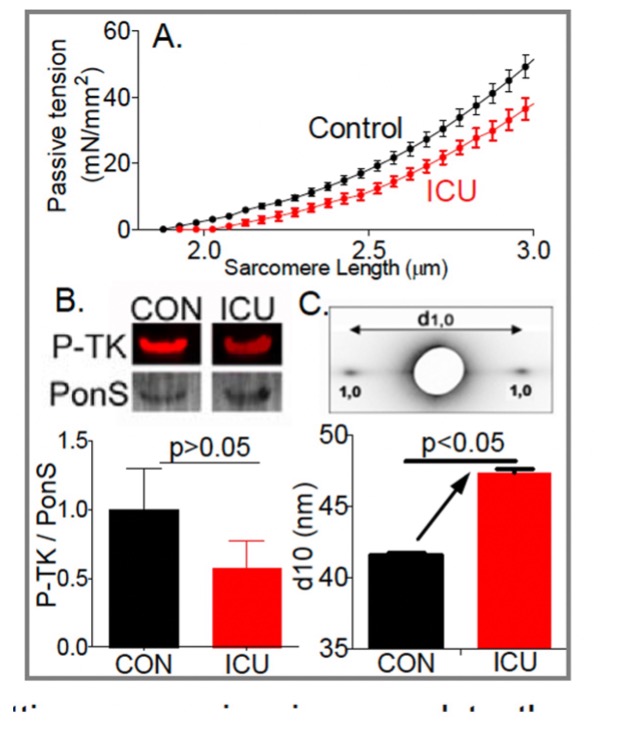
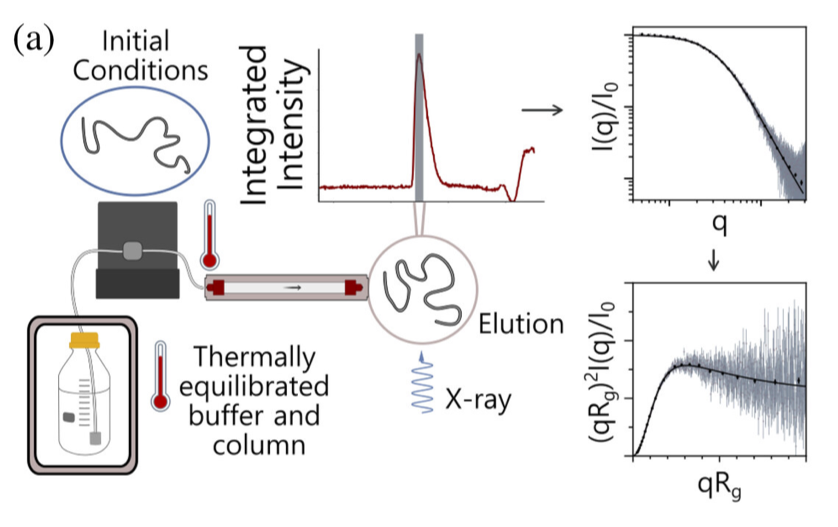
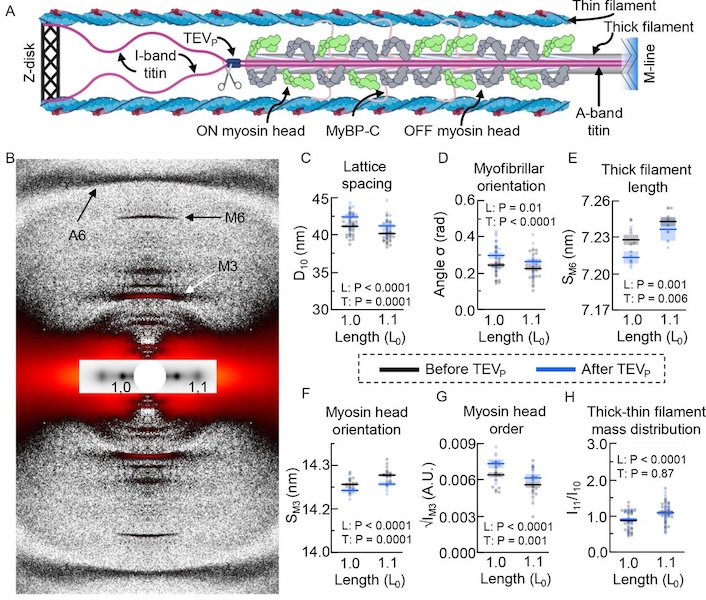
Unraveling folding pathways of dynamic DNA quadruplexes

G-quadruplexes (G4s) are non-B form DNA structures containing genomic regions rich in guanine that sometimes fold into and often act as transcriptional regulators. They are frequently located at regulatory sites including promoters, replication origins and telomeres11. Understanding their folding pathways, while important to a fundamental understanding of form and function, has been challenging. G4 quadruplex folding pathways are distinct from not only proteins but also duplex RNA and DNA12. Previous work characterizing folding pathways using FRET, CD or stopped-flow absorbance suggested a complex, multi-step pathway. Researchers from the University of Louisville, some of whom are frequent BioCAT users, performed continuous-flow mixing time-resolved SAXS experiments to directly structurally characterize, for the first time, early steps in the G4 quadruplex folding pathway.
G4 formation is primarily driven by hydrogen bonding, ion coordination and nucleotide pi-stacking and their resulting structures are stabilized (or destabilized) by a combination of factors including pH, temperature, ionic strength and loop length. Unlike proteins or most duplex nucleic acids, common chaotropic agents such as urea or Guanidinium hydrochloride are not effective at unfolding G-quadruplexes. As such, preliminary equilibrium circular dichroism studies of the systems of interest, two human telomere hybrid structures, were used to identify effective denaturation conditions …
more ...