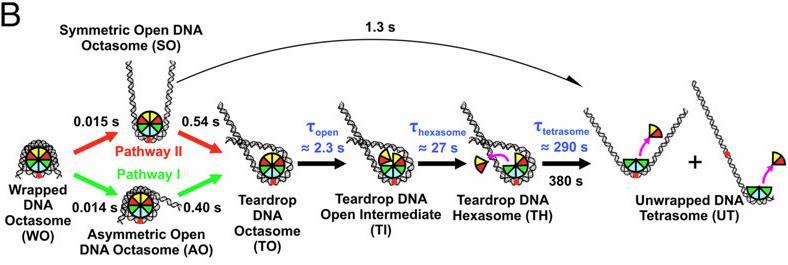
Nucleosomes are protein–DNA structures which eukaryotic organisms use to package and organize DNA inside the nucleus. Nucleosomes need to be disassembled to permit transcription of DNA and reassembled afterwards, hence are an important component of the gene regulation machinery. The nucleosome core particle (NCP) consists of DNA wound around a core of eight histone proteins including two dimers of H2A–H2B histones and an (H3–H4) tetramer that is assembled as a dimer of dimers. A team led by Lois Pollack at Cornell University used time-resolved SAXS at the BioCAT beamline 18ID at Advanced Photon Source and TR-FRET, in collaboration with Lisa Gloss, at Washington State University to study changes in the DNA conformations as a function of the composition of the histone core during salt induced disassembly of NCPs in order to allow identification of kinetic pathways and transient intermediates that show how the sequence of events involving DNA unwrapping and protein dissociation are connected. The investigators found that H2A–H2B histone dimers are released sequentially with an octasome-to-hexasome transition guided by asymmetric unwrapping of the DNA. This work suggests a mechanism for NCP remodeling in which DNA conformation facilitates the reconfiguration of the histone core. This mechanism may be exploited by gene regulatory proteins as a general strategy to exchange or modify one H2A–H2B dimer while simultaneously protecting the other. These dynamic structures provide new insights into the regulation of DNA access during transcription, replication, and repair.
See: Yujie Chen, Joshua M. Tokuda, Traci Topping, Steve P. Meisburger, Suzette A. Pabit, Lisa M. Gloss, and Lois Pollack. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. PNAS January 10, 2017 114:334–339