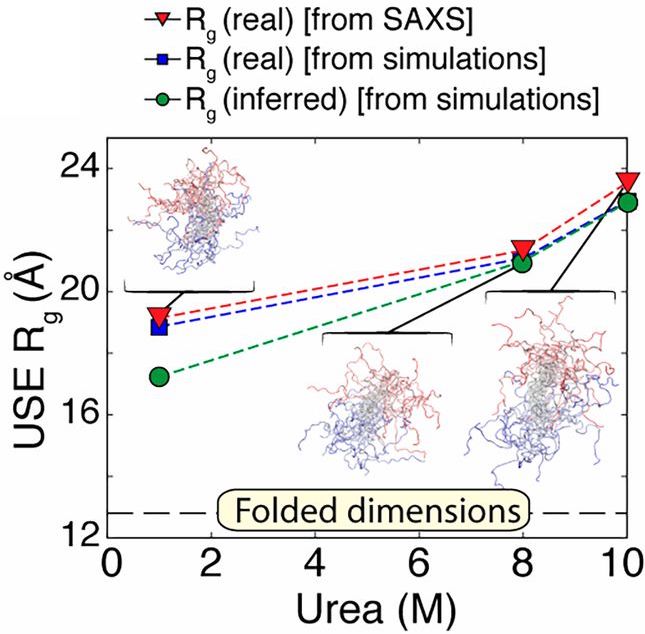
While there is emerging consensus in the protein folding community concerning the behavior of proteins under unfolding conditions, the occurrence of unfolded states under physiological (native) conditions and their propensity to aggregate are the basis of several human pathologies. Valuable insights into these transient species were obtained by taking advantage of the temporal resolution afforded by combining time-resolved fluorescence and continuous (in this case chaotic) flow SAXS (CF-SAXS) with all atom simulations and polymer theory. A group of researchers led by the Raleigh lab (Stony Brook University) used the 59 amino acid N-terminal domain of the ribosomal protein L9 (NTL9), which has a well-studied two state folding mechanism. By introducing FRET pairs several pairwise distance distributions were measured in the unfolded and native conditions in equilibrium and also the unfolded states in native conditions using a continuous flow mixer Interestingly chain contraction as indicated by fluorescence decay was observed well within the dead time of the mixer (~40 µs) showing that chain collapse happens considerably faster than the time-scale required for completion of the folding process (2.5 ms for NTL9). While FRET reports on local interactions, SAXS reports on global shape changes so the two techniques are highly complementary. Data were obtained using SAXS in both equilibrium SAXS and continuous flow time-resolved modes. While the Rgs for the unfolded state at 10 M urea and fully folded state at 1 M urea were determined to be ~ 24 Å and ~ 13 Å respectively using equilibrium measurements, the Rg of the unfolded state at 1 M urea was found to be ~19 Å which represented an approximately 20% contraction. Atomic level information about the different folding states studied using FRET and SAXS was provided by all atom simulations, which, unlike coarse grained models, are able to study sequence-specific interactions and side-chain-specific backbone conformational preferences. These simulations, while confirming the findings from their low-resolution technical counterparts also showed that the contracted but as yet unfolded state of NTL9 found in native conditions is characterized by a mixture of several native-like and nonnative structural features. A novel combination of experimental and theoretical techniques proved to be a powerful tool to explore the always intriguing process of protein folding in increasingly fine detail.
See: Ivan Peran, Alex S. Holehouse, Isaac S. Carrico, Rohit V. Pappu, Osman Bilsel, Daniel P. Raleigh, “Unfolded states under folding conditions accommodate sequence-specific conformational preferences with random coil-like dimensions,” Proc. Natl. Acad. Sci. USA 116 (25), 12301-12310 (2019). DOI: 10.1073/pnas.1818206116