In their continuing endeavor to understand misfolding proteins as part of the etiology of a variety of diseases, the Matthews lab particularly focuses on the different factors that impede a protein’s path from the unfolded state to the global free energy minimum. The complexity of the folding trajectory understandably depends on the size of the protein mostly because of the formation of intermediates many of which often stall the formation of an optimal native conformation.
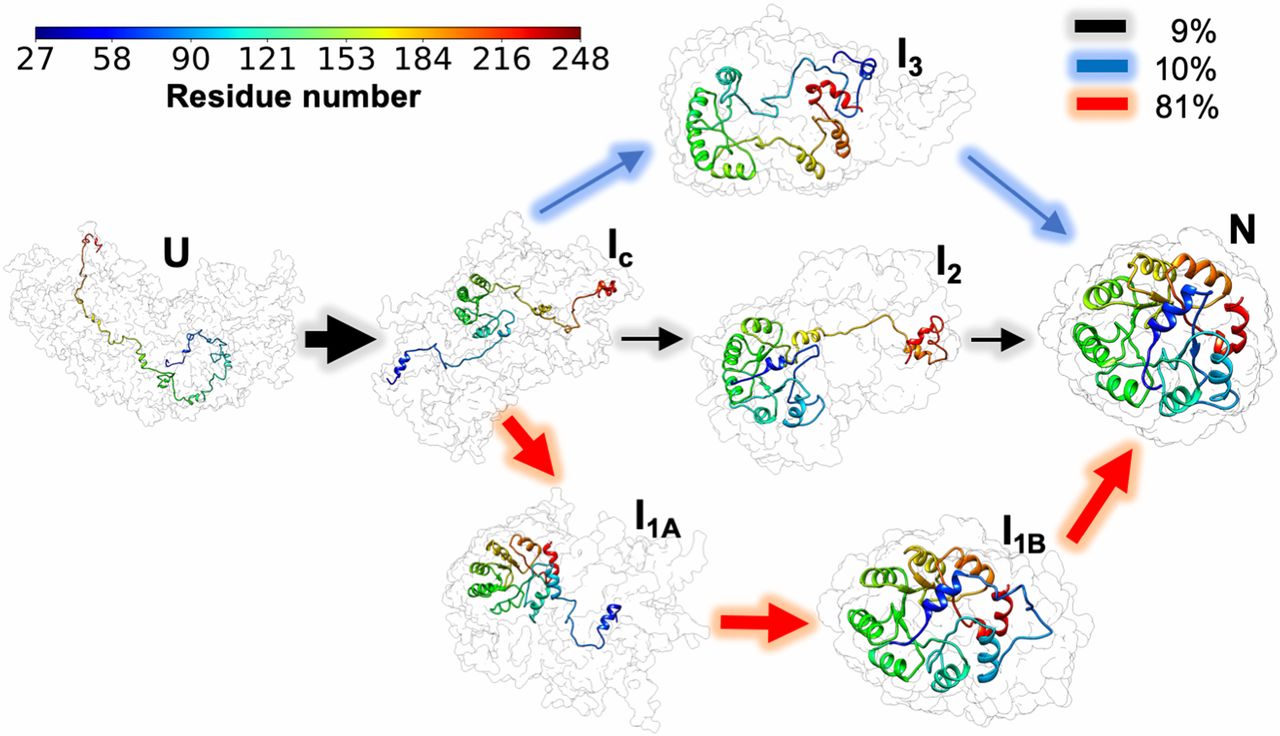
The triosphosphate isomerase (TIM) barrel family of proteins have very well characterized and conserved folding path despite large variations in sequence which make it an ideal group of proteins to obtain widely applicable insights into the folding process. Like other proteins studied before, S.solfataricus indole-3-glycerol phosphate synthase (SsIGPS) a TIM barrel protein goes through a burst phase followed by a relaxation phase and then eventually folds into the native conformation. Mutational and hydrogen exchange experiments have helped characterize the species found in the few millisecond time range of the folding process. In this study, Halloran et al., have used a combination of continuous flow FRET, SAXS and simulations to study the sub-millisecond time window. SsIGPS has 8 βα repeats arranged as a central β barrel encompassed by α helices. In previous studies it has been shown to be kinetically trapped within the first few milliseconds by forming an intermediate IBP which is characterized by a well-protected central (βα)4 segment. This is followed by expansion of protection to a region spanning the (βα)2-6, which subsequently spreads to (βα)1-8 and finally native fold is achieved. The access of previous experimental systems was limited to 10s of milliseconds, which hampered the understanding of the burst phase.
At BioCAT, equilibrium SAXS measurements yielded new structural information concerning the native, and unfolded states which were shown to have a radius of gyration (Rg) of 18 and 46 Å respectively. In addition, a chaotic flow microfluidic mixer was used to acquire time-resolved SAXS data. A ten-fold dilution of SsIGPS in 8 M urea using the mixer led to formation of the IBP state well within the 150 µs dead-time of the mixer and had an Rg of ~26 Å. While the Kratky plot suggested that the IBP is globular, the P(r) curve indicates that there are still unstructured regions and it is significantly different compared to the native conformation. CF-FRET, using more localized distances to monitor conformational changes, further corroborated the formation of IBP during the burst phase even before the first 50 µs. These observations were bolstered by conclusions drawn from coarse grained simulations which showed multiple trajectories that reached intermediates that had different combinations of native and non-native regions. It was also shown that not all trajectories had equal likelihood of achieving the native state. The experimental data were however remarkably consistent with the observations made during simulations and both the approaches strongly supported and further elucidated the models drawn from the hydrogen exchange experiments.
See: Kevin Halloran, Yanming Wang, Karunesh Arora, Srinivas Chakravarthy, Thomas Irving, Osman Bilsel, Charles L. Brooks III, C. Robert Matthews. “Frustration and folding of a TIM barrel protein,” Proc Natl Acad Sci U S A. 2019. 116(33):16378-16383