Estrogen receptor alpha (ERα) plays an important role in the development of various physiological functions; in particular, over 70% of breast cancers are ERα-positive. Although initial treatments that target ERα are often successful, many patients are not responsive or develop resistance so new treatment strategies are urgently needed. ERα is a nuclear hormone receptor that consists of a DNA-binding domain (DBD) and a ligand-binding domain (LBD) that binds the physiological hormone, estrogen, and functions as a hormone-regulated transcription factor. The structure of each domain is known but it is not clear how these two domains communicate with each other. Thus, there is no way to screen for drugs that disrupt this crosstalk. Now, work from a team of researchers from Case Western Reserve University has probed the structure of the complex and its domain interface. The team developed an elegant new approach, based on the new structure, to screen small molecules that target the bridging interface in ERα-positive breast cancer, with the potential for developing new pharmaceuticals to interrupt that communication.
The ERα project was the team’s first application of their iSPOT structural biology platform that combines two complementary synchrotron techniques, small-angle x-ray scattering (SAXS) and hydroxyl radical protein foot-printing, with computational modeling methods. The SAXS provides data on the overall architecture of the complex, while the foot-printing provides an accurate assessment of solvent accessibility and surface topology. Computational docking models are used to fit the combined data for the structure of the complex and to develop testable hypotheses. These methods allowed the researchers achieve medium-to-high-resolution data for the ERα complex and the interface between the two domains.
Hydroxyl radical protein foot-printing was employed at the 5.3.1 x-ray beamline of the ALS at Lawrence Berkeley National Laboratory to compare the solvent accessibility of amino acids on the surface of either the LBD or DBD alone to that of the overall complex to identify residues that appeared to be buried in the complex. Six residues were identified that appear to reside at the domain interface and form two hydrophobic clusters.
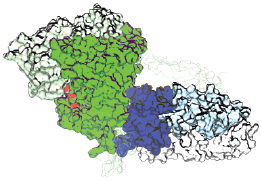
Foot-printing data was then combined with SAXS data from BioCAT to provide overall structural information for computational modeling. The model showed that the ERα complex resembles an L-shaped boot with a short ankle rise and a large foot bed (Fig. 1). The foot bed represents the DBD while the ankle of the boot is the LBD and the “tongue” of the boot is the interface. Utilizing computational methods, researchers demonstrated that the interface is likely to consist of mostly hydrophobic contacts with a medium-sized pocket on the LBD.
To test their modeling predictions, the team mutated residues thought to be important for LDB-DBD interactions and tested them in a reporter assay as a measure of ER-mediated transcription activity. They found that several mutations significantly reduced their transcriptional activity. Interestingly, one mutation found in patients with endometrial cancer was found to possess significantly higher transcriptional activity.
This first test-drive of the iSPOT system appears to have been a great success, providing important information about the unique structural features of ERα and a valuable tool for screening for drugs that might disrupt the interface between the two domains. These drugs could be very useful for treating ER-positive breast cancer using a “burning the bridge” strategy by inactivating ERα activity.
The next step will be to screen the 2000 or so FDA-approved drugs and see if any of them are effective in interrupting domain interactions. In the future, the team hopes to screen new compounds in a larger drug discovery effort.
See: Wei Huang, Yi Peng, Janna Kiselar, Xuan Zhao, Aljawharah Albaqami, Daniel Mendez, Yinghua Chen, Srinivas Chakravarthy, Sayan Gupta, Corie Ralston, Hung-Ying Kao, Mark R. Chance, Sichun Yang, “Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains,” Nat. Commun. 9 (3520), 3520-1-3520-10 (2018). DOI: 10.1038/s41467-018-06034-2