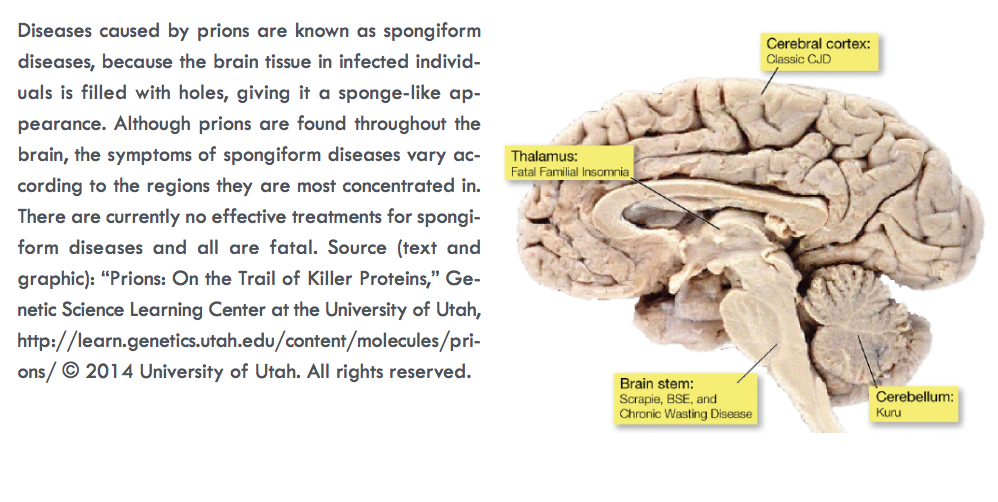
How does one kill an infectious invader that is not technically alive? That question has vexed researchers studying prions—aggregates of aberrantly folded protein that propagate by inducing properly folded proteins to convert into misfolded, disease-associated forms—since these infectious agents were identified as the source of a range of devastating neurological diseases such as transmissible spongiform encephalopathies (TSEs), which include Creutzfeldt-Jakob disease and “mad cow disease.” Prions contain no genetic material, so their structure determines their biological activity. That structure, made rigid and insoluble through aberrant protein folding, also makes prions difficult to combat. Researchers used two U.S. Department of Energy synchrotron light sources including the APS to pry out new structural information about prions, yielding insights into the fundamental mechanisms of prion at a level of complexity that has not been observed in short-peptide amyloids used as prion model systems.
Unlike viruses, bacteria, fungi, and parasites, prions do not spread disease through the transfer of genetic material. They propagate through the insidious conversion of properly folded proteins into deviant forms, which consist of aggregates of amyloids — long, unbranched fibrils assembled into cross-β architecture consisting of one or more tiers of β-strands running perpendicular to the fiber axis, forming β sheets that run the length of the fibril. Notable examples of prion-caused diseases are the aforementioned TSEs, which involve amyloids of the prion protein PrP. Non-PrP amyloids have been implicated in diseases such as Parkinson’s and Alzheimer’s as well as type II diabetes.
Researchers from Vanderbilt University used site-directed mutagenesis together with x-ray fiber diffraction studies to determine the relative importance of various structural features that facilitate deviant folding in a prion model system, the fungal prion HET-s. Their analysis suggests a redundancy of features that enable robust folding and stability even in the face of significant changes to prions’ sequence of constituent amino acids.
Prions’ insolubility and rigid structure make them hard to denature with chemical agents. A possible method to thwart prions is to make them unstable or prevent them from misfolding. In order to develop compounds that exploit these possible mechanisms, it is essential to determine the structural features that give prions the ability to propagate their structures.
To figure out how important each structural element of prions is to the ability to consistently misfold, researchers have looked to a prion model system, the fungal prion HET-s(218-289). Fungal prions are not only incapable of causing diseases, they are in many cases thought to be adaptive. HET-s works well as a model system owing to its highly ordered structure that readily folds into a complex amyloid with two tiers of β-strands.
The researchers used site-directed mutagenesis of amino acid residues to alter the flexible loop sequences, buried polar interactions, salt bridges, and asparagine ladders found in HETs(218–289) in order to probe the relative importance of these features for the formation of β-solenoid structure. They also investigated the cumulative effects of multiple mutations.
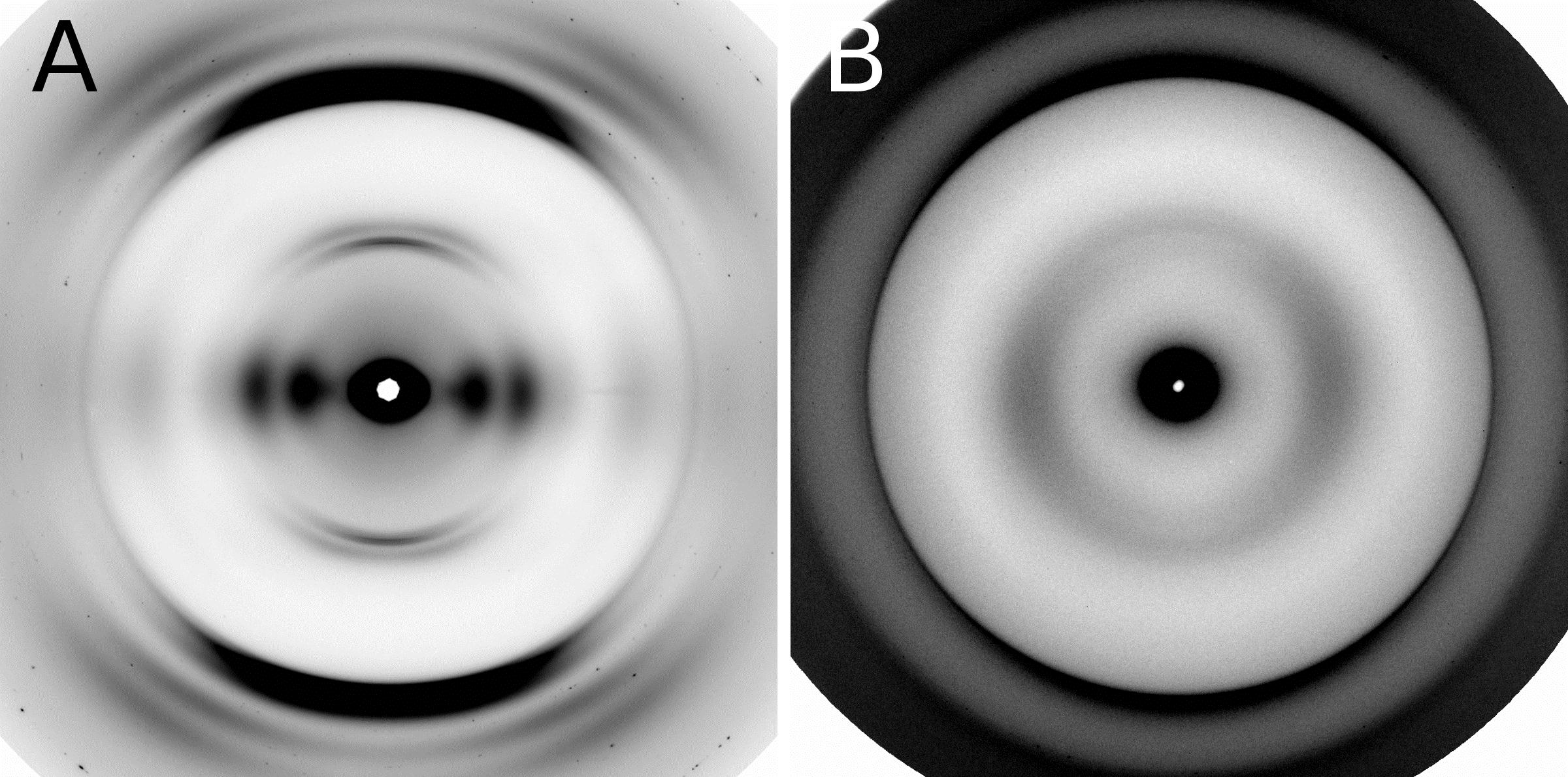
To determine whether the modified structures contained cross-β fibers, the researchers studied the samples with x-ray diffraction (Fig. 1) at the BioCAT beamline 18-ID-D at the APS and at beamline 4-2 at the Stanford Synchrotron Radiation Lightsource at the SLAC National Accelerator Laboratory.
As a final step, mutants that formed β-solenoids were subjected to assays examining the protein denaturation and the kinetics of fibril formation to assess the effects of mutations on the biophysical properties of the β-solenoid structure. The β-solenoid architecture of HET-s(218–289) was found to be surprisingly robust because of the redundancy of important biophysical features. Several mutations significantly impacted fibril nucleation, the rate of fibril formation, and fibril stability, but x-ray fiber diffraction revealed that the β-solenoid structure formed in most cases.
To effectively wipe out the β-solenoid structure required completely rearranging or reversing the long flexible loop connecting the two rungs of the β solenoid or removing both asparagine ladders and all three salt bridges.
In addition, comparing the sequence alignment of HET-s(218-289) to one of its homologs, FgHET-s(218-289) suggests that of these three features, the long flexible loop and the asparagine ladders were most necessary for β-solenoid formation.
The investigation of the fungal prion HET-s(218-289) provides insights into the fundamental mechanisms of prion assembly and propagation of its infectious fold, which is made robust by a complex and diverse array of inter and intramolecular structural features. This level of complexity has not been observed in short-peptide amyloids that have been used as prion model systems.
See: William Wan and Gerald Stubbs, “Fungal prion HET-s as a model for structural complexity and self-propagation in prions,” Proc. Natl. Acad. Sci. USA 111 (14), 5201 (2014). DOI: 10.1073/pnas.1322933111
This work was supported by National Institutes of Health (NIH) Grants AG002132 (to Principal Investigator G.S.; Program Director Stanley Prusiner) and F31-AG040947 (to W.W.). Bio-CAT is supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the NIH. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Adapted from an APS press release by Chris Palmer.