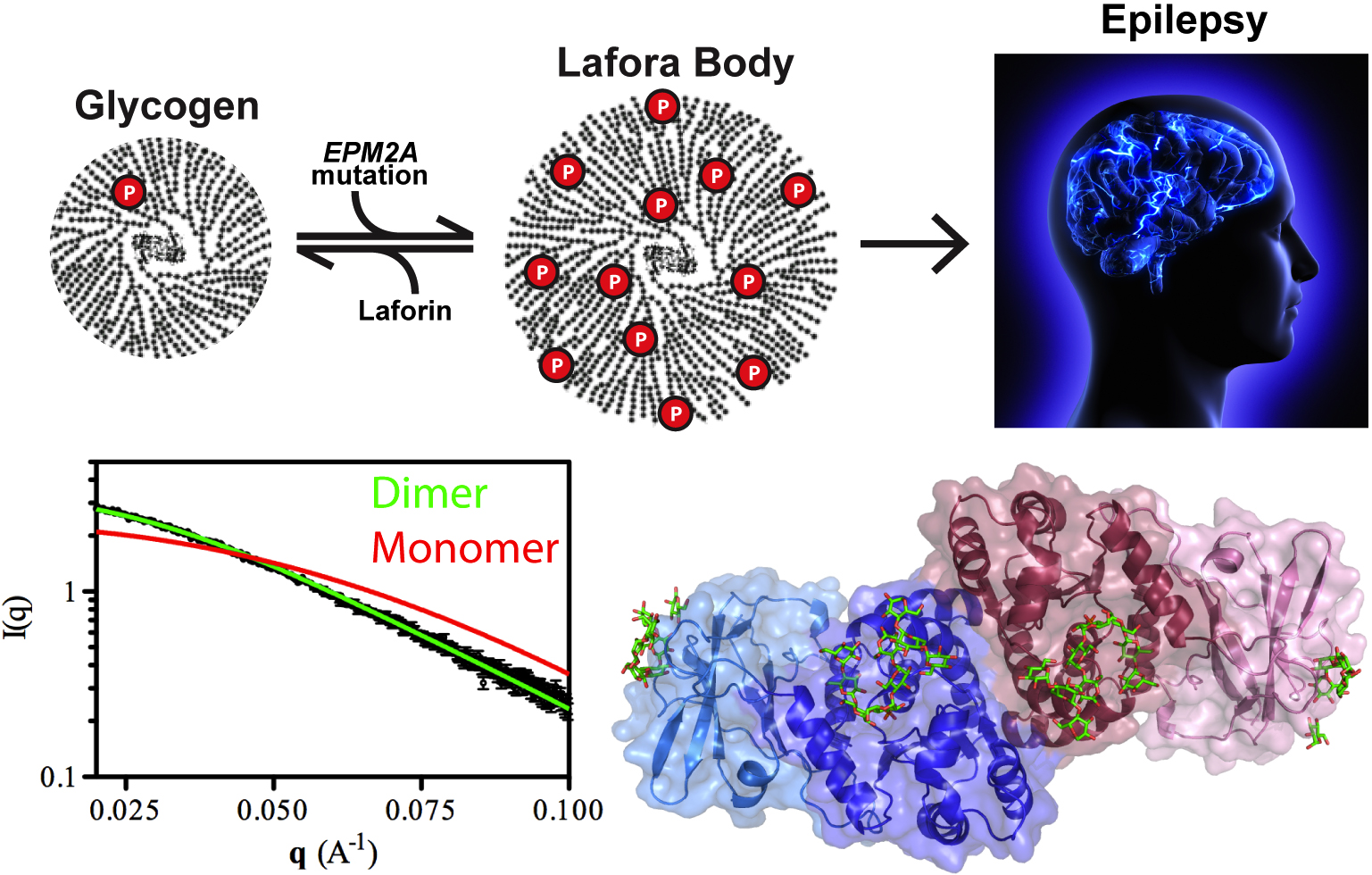
Our cells are brilliant biochemists that solve all sorts of chemistry problems under difficult conditions. They speed up slow reactions by orders of magnitude, stuff miles of DNA into tiny spaces, and carefully balance the solubility of different kinds of molecules in a jam-packed cellular solution. A good example is the storage of glucose energy in cells in the form of glycogen. Cells can store up to 55,000 glucose units in water-soluble spheres of branched, polymeric glycogen. This provides ready energy for rapid response to cellular needs but also must be managed carefully because too much glycogen accumulation can activate programmed cell death. This is especially true of neurons, which consume large amounts of glucose but are particularly sensitive to glycogen build-up. One example of what can happen when this basic metabolic process goes awry is observed in Lafora disease, a devastating fatal epilepsy in which mutations in a single key enzyme result in the formation of insoluble glucan inclusion bodies that cause neuronal death. Research conducted at two x-ray beamlines at the U.S. Department of Energy’s Advanced Photon Source (APS), an Office of Science user facility at Argonne solved the structure of the enzyme responsible, the laforin glucan phosphatase. The work has provided important insights into both the basis of Lafora disease and normal glycogen metabolism.
Patients with Lafora disease develop epilepsy in their second decade and the severity of their seizures progressively worsens until their death in 5-10 years. A number of mutations have been discovered in the laforin phosphatase that are responsible for the glycogen solubility defect and the eventual formation of the insoluble Lafora bodies but developing targeted treatment requires deeper understanding of how the mutations affect the activity of the laforin protein. Utilizing the SER-CAT 22-ID-D beamline at the APS, the research team from the University of Kentucky; the University of California, San Diego; the University of Notre Dame; the Illinois Institute of Technology; and the lnstituto de Biomedicina de Valencia solved the structure of laforin (2.4 Å) in complex with catalytic products maltohexose and phosphate bound to the laforin dual-specificity phosphatase catalytic domain (DSP) and carbohydrate binding domain (CBM) in a specific orientation (Fig. 1). Further, the dimeric architecture of the protein in solution was defined using size exclusion chromatography-small angle x-ray scattering experiments at BioCAT, resolving an important outstanding question in the field.
When researchers turned to the question of how Lafora disease mutations affect the activity of the laforin enzyme, they found that, in addition to harboring mutations that affect the active site of the DSP domain, Lafora disease mutations could also be found in the CBM domain and in the interface between the DSP and CBM domains. Mutations in the DSP domain affected catalytic activity of the enzyme while those in the CBM domain were found to reduce glycogen binding and overall protein stability. Those in the domain interface diminished enzymatic activity but not glycogen binding. These data suggest that the unique tetramodular orientation of the domains is critical to laforin function. Also, unlike its enzymatic relative in plants that breaks down starch, laforin binds to multiple maltohexose chains in a particular orientation instead of one long carbohydrate along the length of the DSP and CBM domains. This leaves open an important question regarding how laforin acts on branch points in the glycogen polymer and will be the focus of the team’s future work.
Recent work in the field of epilepsy has suggested that, rather than being a defect of neuronal firing, epilepsy may be a metabolic disorder similar to diabetes. In fact, this hypothesis is also being proposed for Alzheimer’s disease and glycogen metabolic defects have been linked to neuronal decline in aging, glycogen storage diseases, diabetes, cancer, and some cardiac syndromes. This work provides important insights into basic metabolic processes that have critical underpinnings in many areas.
Understanding how laforin regulates the formation of glycogen spheres and how the delicate balance of solubility versus fatal neuronal damage is maintained will be important to development of treatments for Lafora disease and potentially many others.
See: Madushi Raththagala, M. Kathryn Brewer, Matthew W. Parker, Amanda R. Sherwood, Brian K. Wong, Simon Hsu, Travis M. Bridges, Bradley C. Paasch, Lance M. Hellman, Satrio Husodo, David A. Meekins, Adam 0. Taylor, Benjamin D. Turner, Kyle D. Auger, Vikas V. Dukhande, Srinivas Chakravarthy, Pascual Sanz, Virgil L. Woods, Jr., Sheng Li, Craig W. Vander Kooi, and Matthew S. Gentry, “Structural Mechanism of Laforin Function in Glycogen Dephosphorylation and Lafora Disease,” Mol. Cell 57, 261 (January 22, 2015). DOI: 10.107 6/j.molce1.2014.11.020