The molecular interactions between the proteins myosin and actin that generate force during muscle contraction are some of the most well-studied molecular interactions in biology. However, there are some congenital skeletal muscle disorders and types of heart failure where relaxation of the muscle, rather than the force generation part of the cycle, appears to be the problem, and there are currently no available treatments that affect relaxation specifically. A more detailed understanding of the dynamics of the relaxation process could help in the development of treatments that maintain or increase force generation while repairing defects in relaxation. Recent work conducted at BioCAT used a unique transgenic mouse model, time-resolved small-angle x-ray diffraction, and molecular dynamics simulations to discover more about how myosin and actin interact during skeletal muscle relaxation. This research, published in the Journal of Physiology, demonstrates that this type of small-angle x-ray analysis may be of great value for uncovering the information needed to identify new treatments for neuromuscular disorders associated with impaired muscle relaxation kinetics.
The protein-protein interactions between myosin and actin to generate skeletal muscle contraction forces involve a cycle that starts with myosin thick-filament binding to actin thin filaments to form cross bridges. This is followed by conformational changes in the myosin that pull muscle filaments toward each other and then release the filament to enter relaxation. The process is dependent on many other proteins, calcium, and, like so many other cellular processes, the hydrolysis of adenosine triphosphate (ATP). In the laboratory, researchers have shown that other nucleotides can affect the dynamics of myosin cycling, including the naturally occurring 2-deoxy-ATP (dATP) that is a myosin activator that enhances the rate and magnitude of force development. However, the reason it attracted the interest of this research team is that the effects of dATP on contraction kinetics also offer an opportunity to study the relaxation process in more detail.
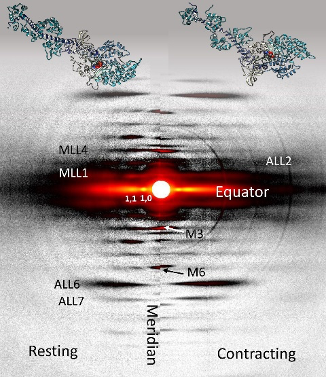
For the muscle experiments, the team of researchers from the Illinois Institute of Technology, the University of Washington, and Cornell University was able to take advantage of a transgenic mouse model in which the expression levels of an enzyme regulating the formation of dATP are elevated in skeletal muscle tissue, increasing the amount of dATP in the muscle 25-fold to a total of 1%-2% of total ATP. Comparison of time-resolved small-angle x-ray diffraction data from non-transgenic mice to the transgenic mouse model tissue obtained at BioCAT allowed the team to make some discoveries about the effects of dATP on skeletal muscle relaxation (Fig. 1).
The main observations from comparison of small-angle x-ray diffraction data for the two muscles were that, although there were no differences in the kinetics of force development compared to the wildtype, the myosin in the dATP-enhanced muscles was closer to the actin, took more time for the force to drop, had a longer half-time for relaxation, and returned to the resting state more quickly. The high time resolution of the high-brightness APS x-rays combined with the x-ray diffraction technique allowed the team to distinguish the nuanced effects of dATP by capturing the prolonged decay in force relative to the wildtype muscle as well as recording the faster restoration of the resting state, discrete steps that could easily have been missed with less sensitive techniques.
These findings led the team to ask what is different about myosin in the presence of dATP? For this they turned to molecular dynamics simulations. Modeling of myosin bound to dATP showed that it adopted more conformations with dATP than with ATP and that it particularly favored an extended conformation that may be more accessible to actin. In addition, solvent accessible surface area (SASA) analysis suggested that dATP increases the surface area of actin binding regions, increasing the exposure of positively charged residues near the negatively charged actin and potentially explaining the observation that myosin in the dATP muscles was closer to the actin filaments.
See: Weikang Ma, Matthew Childers, Jason Murray, Farid Moussavi-Harami, Henry Gong, Robert Weiss, Valerie Daggett, Thomas Irving, and Michael Regnier, “Myosin dynamics during relaxation in mouse soleus muscle and modulation by 2’-deoxy-ATP,” J. Physiol. 598(22), 5165 (2020). DOI: 10.1113/JP280402