TAP binding protein, related (TAPbPr), a novel protein chaperone, plays a role in loading peptides onto major histocompatibility class i (mhc i) molecules during the process of immune surveillance. However, until now, how TAPbPr functions in antigen presentation has been unknown. Researchers investigated the biochemical function of TAPbPr, comparing it with tapasin, another chaperone with a similar protein sequence. Using direct binding studies, they demonstrated that TAPbPr functions as a peptide editor, and binds mhc i molecules that are either peptide-free or complexed with low affinity peptides. Because macromolecular crystallography has so far proven unsuccessful for obtaining high-resolution structural information on TAPbPr, the researchers instead collected small-angle x-ray scattering (SAxS) data at the APS. This allowed them to confirm the structural similarities between TAPbPr and tapasin. The results of this study could lead to ways to modulate peptide loading in vaccine design, improving T-cell recognition.
TAPbPr is involved in the intracellular loading of peptides onto mhc i molecules. Mhc i molecules are important components of the immune system and are expressed on the surface of most cells in all species of jawed vertebrates. Many variants of mhc i molecules exist within a population, and they bind and display peptides on their surface: in healthy cells, they display peptide fragments derived from biosynthetic pathways—typically from the cell’s own “housekeeping” proteins—which do not stimulate an immune response. However, if a cell is infected by a microorganism, or if a cell is a dysregulated tumor cell, mhc i molecules bind and display peptide fragments of the microbe or the tumor. These mhc i–antigen complexes serve as flags on the cell surface that are recognized by T cell receptors on cytotoxic T lymphocytes.
TAPbPr shares approximately 20% of its protein sequence with tapasin, another well-studied chaperone. Tapasin is part of a complex protein machine called the “peptide loading complex,” which is the major loader of peptides onto mhc i molecules. However, how TAPbPr functions in peptide loading has been unclear.
The researchers in this study, from the National Institutes of Health, the U.S. Food and Drug Administration, and the University of Oklahoma Health Sciences Center conducted studies to characterize the biochemical function of TAPbPr. They wanted to better understand how TAPbPr interacts with mhc i molecules to facilitate peptide loading. They also wanted to determine the structure of TAPbPr, and to compare it with the structure of tapasin, which is already known.
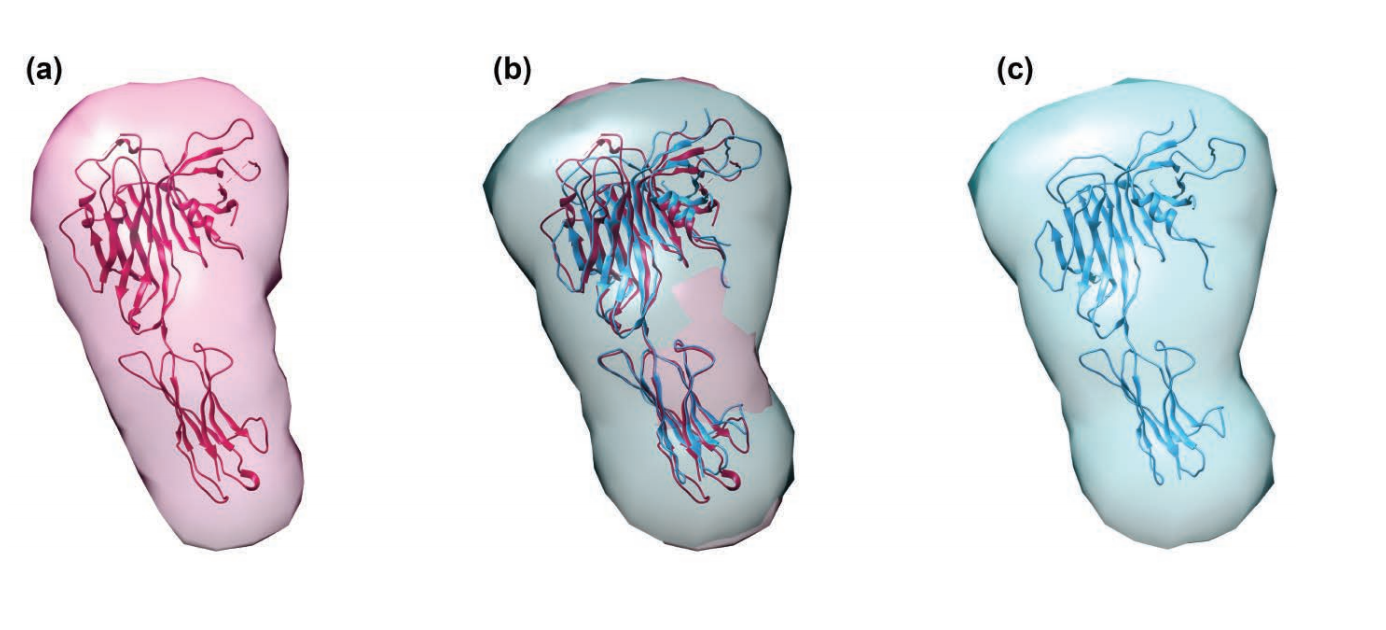
The researchers engineered recombinant TAPbPr for expression in an insect cell system. They investigated the interaction of TAPbPr with mhc i molecules, by using direct binding studies including analytical ultracentrifugation, surface plasmon resonance, and fluorescence polarization assays. These studies showed that TAPbPr functions as a peptide editor and binds mhc i molecules that are either complexed with low affinity peptides or emptied of photolabile peptides by exposure to ultraviolet light. As noted, researchers have so far been unsuccessful in using crystallography to obtain high-resolution structural information on TAPbPr. Instead of using crystallography, the researchers in this study took a different approach to determining and comparing the structural shapes of purified TAPbPr molecules and the related tapasin molecule. By collecting SAXS data at the BioCAT 18-id-d x-ray beamline at the APS, they were able to calculate a low-resolution envelope for both tapasin (fig. 1a) and TAPbPr (fig. 1c). They were also able to fit the x-ray structures of tapasin and a molecular model of TAPbPr into their respective SAXS-determined low-resolution envelopes (fig. 1).
In this way, the researchers identified the similarities between the two molecules that provide the foundation for investigating the steps of peptide loading. The results of this research therefore have potential to guide future studies to modulate peptide loading for T-cell recognition, as well as in designing peptide-based vaccines.
See: Giora I. Morozov, Huaying Zhao, Michael G. Mage, Lisa F. Boyd, Jiansheng Jiang, Michael A. Dolan, Ramesh Venna, Michael A. Norcross, Curtis P. McMurtrey, William Hildebrand, Peter Schuck, Kannan Natarajan, and David h. Margulies, “Interaction of TAPbPr, a tapasin homolog, with mhc-i molecules promotes peptide editing,” Proc. Nat. Acad. Sci. USA 113(8), e1006 (February 23, 2016). doi: 10.1073/pnas.1519894113.
SAxS research was supported by grant 9 P41 gm103622 from the National Institute of General Medical Sciences of the National Institutes of Health. The research was supported by the intramural research programs of the National Institute of Allergy and Infectious Diseases (Project Ai0000394) and National Institute of Biomedical Imaging and Bioengineering (Project eb000008)/National Unstitutes of Health and the Center for Drug Evaluation and Review/Federal Drug Administration (Project 1617). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National laboratory under contract no. de-Ac02-06ch11357.
Adapted from an APS press release by Nicola Parry.