It is hard to believe that a single protein can be responsible for the damage inflicted by diseases such as human Creutzfeldt-Jakob and bovine spongiform encephalopathy (Mad Cow Disease). Yet the implicated protein, known as a prion and only about 200 amino acids long, can initiate and propagate a disease cycle just by changing its shape. Prions are amyloids, which are misfolded proteins now implicated in numerous diseases. Studying the prion diseases has required patience and fortitude because of disorder and insolubility of the prion samples. Aided by four U.S. Department of Energy x-ray beamlines (BioCARS 14-BM and BioCAT 18-ID at the Advanced Photon Source at Argonne, the 4-2 beamline at the Stanford Synchrotron Radiation Laboratory, and 12.3.1 at the Advanced Light Source), a collaborative research team made up of scientists from Vanderbilt University and the University of California, San Francisco have achieved a significant advance in our understanding of the infectious power of the prion protein.
Gerald Stubbs, who led the research at Vanderbilt University, says that prions and other misfolded proteins, such as those associated with Alzheimer’s disease, represent “both a fascinating problem in protein folding and an excellent application of fiber diffraction.” So when Stanley Prusiner, who received the Nobel Prize in Physiology or Medicine in 1997 for his work on prions, called in 2003 and asked Stubbs to collaborate on structural studies of prions, Stubbs said he was “delighted.”
Previous work had definitively shown that a particular conformation, or isoform, of the mammalian prion protein was responsible for causing the central nervous system (CNS) diseases now known as prion diseases: Creutzfeldt-Jakob, bovine spongiform encephalopathy, ovine scrapie, and other mammalian CNS disorders.
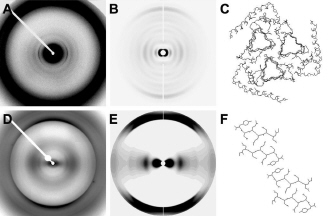
The team used the Advanced Photon Source and Stanford Synchrotron Radiation Laboratory facilities to obtain diffraction patterns from infectious prions, compared them with diffraction patterns from genetically engineered prions, and discovered important differences, both in structure—as determined by diffraction studies, and in heterogeneity—as determined by electron microscopy (see the figure above). Both types of samples exhibit cross-β diffraction, which is indicative of amyloid structure. The most prominent difference was equatorial intensity indicative of stacked β-sheets, which is present in the recombinant samples but missing from the brain-derived prions. Some of the patterns from infectious prions produced data consistent with a repeating unit of 4 β strands, which is consistent with the group’s earlier model derived from electron crystallography.
Discovering the structure to be cross-β was, said Stubbs, “not surprising.” He points out that, for many years, researchers had proposed that the prion structure was a cross-β structure, and “the most convincing models from our collaborators and other groups all incorporated this feature. It took many years to prove this, however. That was only done with our work.” Stubbs adds: “Recent work is showing that biologically significant amyloids come in a variety of structures, all cross-β, but with the β-strands connected in very different ways. As in so many other places, nature is far more inventive than we are.”
On an interesting side note, the research team found that the synthetic prions recovered from mice inoculated with recombinant prions were more like the naturally occurring prion proteins than the recombinant ones. The investigators put forth several hypotheses that would explain the observed structural differences, including the intriguing idea that only some of the recombinant prion proteins have a conformation that would make them infectious or that some difference in the recombinant protein is inhibiting its transformation to an infectious conformation.
Though more work needs to be done to pinpoint the exact structural changes that make the prion infectious, the present work sets the stage for that next set of experiments. Stubbs says that the team is “particularly interested in looking at strains of prions in which the molecules are slightly differently folded, with significant consequences for infectivity, symptoms, and even in rare cases the ability to be transmitted from one species to another.” Transmission across species is, according to Stubbs, what makes the question of strain structure “one of the most interesting in prion research at present.”
By elucidating and underscoring the differences between the brain-derived infectious form and the recombinant forms of the prion proteins, the present work provides bright beacons on the path to understanding the infectious power of the prion protein.
See: Holger Wille,, Wen Bian, Michele McDonald, Amy Kendall, David W. Colby, Lillian Bloch, Julian Ollesch, Alexander L. Borovinskiy, Fred E. Cohen, Stanley B. Prusiner, and Gerald Stubbs, “Natural and synthetic prion structure from X-ray fiber diffraction,” Proc. Natl. Acad. Sci. USA 106(40), 16990 (2009). DOI: 10.1073_pnas.0909006106
This work was supported by National Institutes of Health (NIH) grants NS064, AG010770, and AG02132; the Fairchild Foundation; the G. Harold and Leila Y. Mathers Foundation; and a Jane Coffin Childs postdoctoral fellowship (to D.W.C). Fiber diffraction data analysis software from FiberNet (www.fiberdiffraction.org), which is supported by National Science Foundation grant MCB-0234001. Diffraction data were collected at BioCARS under the BioCAT/BioCARS collaborative agreement; preliminary data were collected at beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. The SSRL and ALS are supported by the U.S. Department of Energy; SSRL beamline 4-2, BioCAT, and BioCARS are also supported by the NIH National Center for Research Resources.
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The Stanford Synchrotron Radiation Lightsource is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences.
Based on an APS press release by Mona Mort.