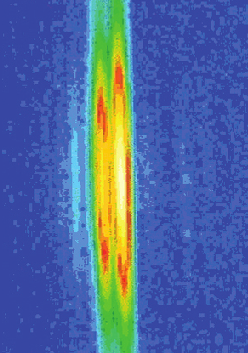
Any time we move our arms, billions of protein molecules work in unison to contract one muscle while letting another muscle relax. The mechanical details of muscle contraction have been known for a long time, deduced from experiments with frog legs, reconstruction of the proteins involved in crystallographic studies, and many other kinds of research. But no one has ever taken such detailed time-lapse molecular pictures of the proteins at work within living muscle. Researchers from Brandeis University, the University of Florence, and Illinois Institute of Technology investigated whether the muscle-contracting proteins work the same way inside muscles as they do inside laboratories. But muscle cells are full of water and the proteins don’t line up quite as regularly as they would purified in crystals. So rather than spend 30 to 40 hours collecting data with a typical x-ray tube, the researchers turned to the BioCAT beamline 18-ID at the APS. With a million times more intensity than a “home source” x-ray tube—and the ability to focus the x-ray beam into a narrow line—the team got a detailed view of the proteins at work (Fig. 1). Not only did they measure the degree of movement of the proteins, but they saw that the proteins synchronized themselves in living muscle much better than anticipated.
Although muscles look like a long single unit to the eye, they are actually made up of small segments called sarcomeres, linked together along the length of a single cell. Every sarcomere contracts at about the same time, shortening the overall length of the muscle. Proteins are the workhorses behind the contraction. One row of proteins—known as actin—forms a kind of stiff track along which other proteins—called myosin—attach. Myosin molecules have long handles that wrap around each other to form a connected, rigid backbone. From this backbone rise lever arms, which are topped by flexible heads that grab onto the actin track. As the lever arm swings on its pivot, the heads pull the actin filaments, shortening the sarcomere.
To view this activity in a living muscle, the team first stimulated the thigh muscle of a Rana pipiens (commonly known as the Northern Leopard Frog) to make the muscle contract, and then kept the muscle tense. They then released the muscle quickly and briefly—within a millisecond—so that it shortened by about 1% of its length. At the same time, the extreme brilliance of APS x-ray beams allowed the researchers to take “snapshots” of the proteins. From the scattering of x-rays by the actin and myosin, the team could reconstruct the positions of the different parts of the proteins: the track, the heads, the handles and lever arms.
The myosin heads start out almost perpendicular to the actin track: about 10º to 15º off, with a group of myosin heads every 14 nm. Then, when the muscle shortens, the arms swing a full 60º through the perpendicular axis, moving the heads a total distance of 10 nm.
In addition, the more in-sync the myosin proteins moved with each other, the clearer the data were that the team collected. The researchers expected the myosin heads to be at a variety of angles. But the lever arms only differed by a few tens of degrees from each other. Because most other data on actin and myosin have been collected from purified muscle proteins, this work reveals a more accurate image of what’s going on in active, intact muscles.
See: Hugh Huxley, Massimo Reconditi, Alex Stewart, and Tom Irving, “X-ray interference studies of crossbridge action in muscle contraction: evidence from quick releases,” J. Mol. Biol. 363, 743 (May 2006). DOI:10.1016/j.jmb.2006.08.075
BioCAT is a National Institutes of Health-supported Research Center RR- 08630. H.E.H. received support during part of this work from NIH Grant no. AR43733. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W31-109-ENG-38.
Based on an APS press release by Mary Beckman.