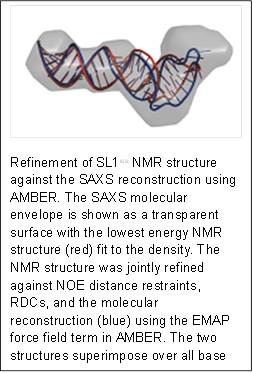
Human immunodeficiency virus type 1 (HIV-1) requires controlled synthesis of its protein complement for persistent infection and successful virion production. Genome expression is tightly regulated at the levels of transcription, splicing, mRNA nuclear export, and translation. RNA polymerase II-dependent transcription yields a 9-kilobase (kb) polycistronic transcript that undergoes multiple rounds of alternative splicing to produce upward of 100 different viral mRNAs that recruit antagonistic host RNA-binding proteins. Thus, HIV-1 splicing pathways are essential components of the viral replication cycle and represent new targets for therapeutic intervention. Because HIV-1 splicing depends on protein-RNA interactions, it is important to know the tertiary structures surrounding the splice sites. Herein, we present the NMR solution structure of the phylogenetically conserved ISS stem loop. ISS adopts a stable structure consisting of conserved UG wobble pairs, a folded 2X2 (GU/UA) internal loop, a UU bulge, and a flexible AGUGA apical loop. SEC-SAXS data collected at BioCAT was used to confirm the general features of the NMR structure and by refining the NMR structure against the SAXS data, obtain a better structure than that of NRM alone. Collectively, this work provides additional insights into how HIV-1 uses a conserved RNA structure to commandeer a host RNA-binding protein.
This project made excellent use of the superior quality SAXS data obtainable with Size Exclusion Chromatography SAXS at BioCAT
Citation: Niyati Jain, Christopher E. Morgan, Brittany D. Rife, Marco Salemi, Blanton S. Tolbert, “Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1,” J. Biol. Chem. 291 (5), 2331-2344 (2016). DOI: 10.1074/jbc.M115.674564 PMCID:PMC4732216