For many proteins, the ability to change shape is essential for their proper functioning within cells. One longstanding question concerns the process proteins follow when shifting from an unfolded to a three-dimensional globular form. Most previous studies have supported the idea that when an unfolded protein is exposed to native conditions (i.e., to an environment favoring its typically fully-folded, physiological form) a continuous unfolded-to-semi-folded collapse ensues. However, other studies suggest that there is an energetic bottleneck to this step that renders it an all-or-none transition. To resolve the issue, researchers from the University of Massachusetts Medical School and the Illinois Institute of Technology probed the folding stages of cytochrome c, an archetype for protein folding behavior. Its microsecond-scale folding dynamics were unambiguously characterized with Förster resonance energy transfer (FRET) complimented by small angle x-ray scattering (SAXS) carried out at the APS. The SAXS and FRET measurements refuted the conventional view that cytochrome c folding proceeds immediately and smoothly when exposed to native conditions; instead, subpopulations of unfolded and semi-folded protein were observed to coexist during brief intervals. These results provide fundamental insights for biochemistry, as well as for human diseases characterized by protein misfolding, including Parkinson’s, Huntington’s, Alzheimer’s, and lysosomal storage diseases.
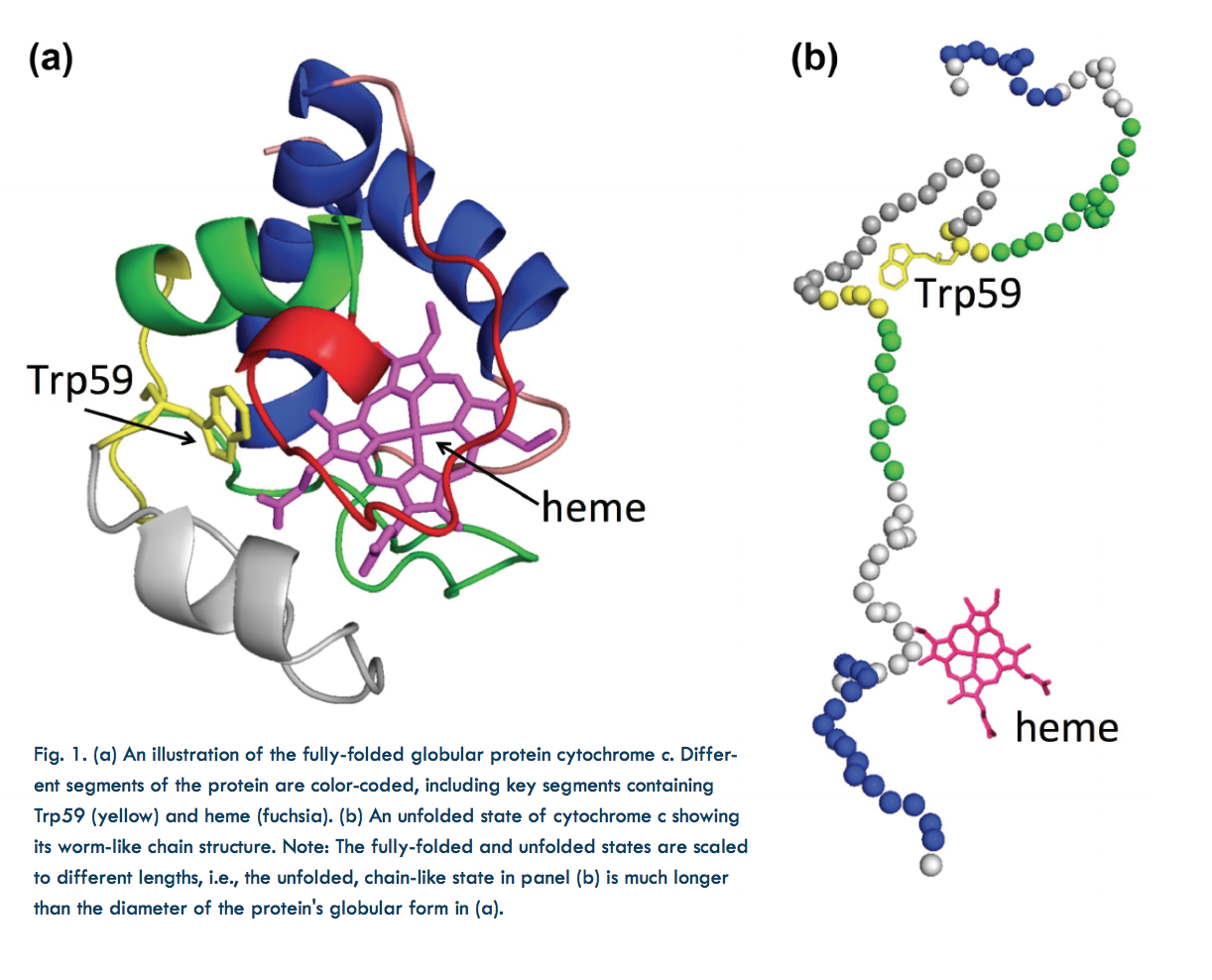
In the laboratory, protein structure can be altered by varying the temperature, chemical environment, etc. For instance, the shapes of proteins suspended in solution can be changed by varying the concentration of a chemical denaturant. In this study, guanidinium hydrochloride (GdnHCl) was used as a denaturant to uncoil cytochrome c, a small, globular, iron-containing protein (hemeprotein). Figure 1a shows its globule form, Fig. 1b an unfolded state.
Cytochrome c molecules were initially suspended in a highly-concentrated solution of GdnHCl, which relaxed some of the protein globules into a chain-like (unfolded) form. To start the folding process, the protein chains were suddenly exposed to a near-native state (i.e., to an environment with little denaturant) using microfluidics techniques that allowed extreme dilution of the GdnHCl-concentrated solution in just 25-30 µs.
Previous experiments capable of resolving early (<100 µs) folding dynamics were unable to differentiate between partially-folded and unfolded forms mixed together. By contrast, the FRET and SAXS techniques were capable of detecting the presence of both folded and unfolded subpopulations immediately following dilution of the GdnHCl denaturant.
FRET employed rapid laser pulses to excite the Trp59 chromophore (a molecular segment that gives the protein its color). Due to their close proximity (several nanometers) and similar electronic energy levels (the “resonance” in FRET), energy could pass from the Trp59 to the heme molecule (called the “donor” and “acceptor,” respectively). This is a quantum mechanical, non-radiative process (i.e., no actual photon is transferred). Energy accepted by the heme molecule in this process was dissipated. However, sometimes the excited Trp59 molecule re-emitted the energy as a photon, which could then be detected by a sensor. The time between a laser pulse and Trp59 photon emission was measured. Because this interval varied according to the Trp59- to-heme distance (which indicates protein shape) the proportions of any unfolded and partially-folded states could be calculated.
The FRET results revealed that following denaturant dilution, certain subpopulations dominated during particular time intervals: within the first 30 µs after dilution, the unfolded state (denoted as “state 1” for convenience) dominated, accompanied by a small proportion of partially-folded states. After 30 µs, a transition to a partially-folded “state 2” occurred, followed still later by dominance of partially-folded “state 3.” These discrete jumps in subpopulation ratios, which occurred as the initially unfolded protein transitioned to the globular form, are referred to as “barrier-limited chain collapse.”
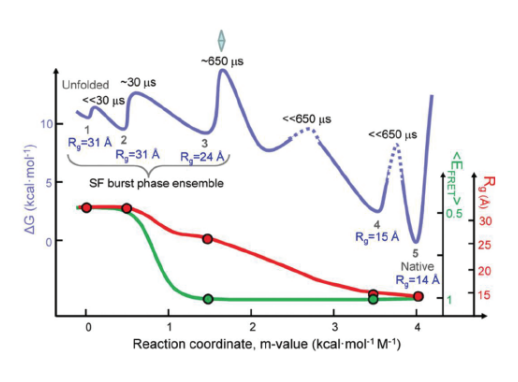
The SAXS experiments, which were carried out at the BioCAT 18-ID-D beamline at the APS were performed as an independent check on the FRET results. SAXS measured a parameter called the radius of gyration (Rg). This parameter quantified the spatial distribution of all of the molecular components of cytochrome c, thereby indicating the degree of folding. Figure 2 depicts folding dynamics as measured by a combination of the SAXS and FRET methods. The top curve in the figure gives a sense of the barrier limits seen at various stages of the folding process. Each phase in the dynamic folding process exhibits a particular Rg, which in turn indicates a particular structural state (a smaller Rg implies greater protein compaction).
The SAXS and FRET results highlight the importance of employing techniques that can detect subpopulations in folding reactions. Moreover, firmly establishing the existence of barrier-limited protein folding will propel the search for the physical source of this behavior.
See: Sagar V. Kathuria, Can Kayatekin, Raul Barrea, Elena Kondrashkina, Rita Graceffa, Liang Guo, R. Paul Nobrega, Srinivas Chakravarthy, C. Robert Matthews, Thomas C. Irving, and Osman Bilsel, “Microsecond Barrier-Limited Chain Collapse Observed by Time-Resolved FRET and SAXS,” J. Mol. Biol. 426, 9 (May 1, 2014). DOI: 10.1016/j.jmb.2014.02.020
This work was supported by National Institutes of Health (NIH) grants GM23303 and GM54836 and National Science Foundation grants MCB0327504 and MCB1121942, and by grants from the National Center for Research Resources (2P41RR008630-17) and the National Institute of General Medical Sciences (9 P41 GM103622-17) from the NIH. BioCAT is supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Adapted from an APS press release by Philip Koth.